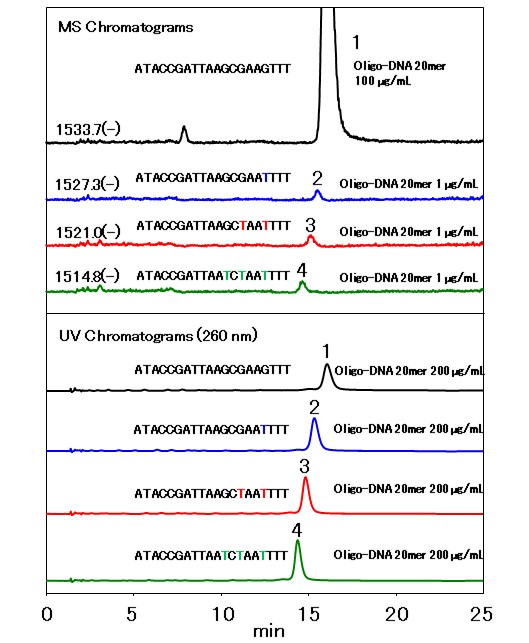
In R&D and QC of nucleic acid drugs such as antisense oligonucleotides (ASO) require development of analytical methods that separate the target synthetic oligonucleotide from its impurities as much as possible. In this application, four compounds were analyzed using HILICpak VN-50 2D, a polymer-based HILIC column. The four compounds were 20mer synthetic oligo-DNAs: one with the target base sequence and its three analogs with 1 to 3 base alterations. By having different bases make their hydrophilicities different from each other. Four synthetic oligo-DNAs were separated by HILIC mode using this hydrophilicity differences. The application developed here does not require a use of ion-pairing reagent nor highly concentrated salt in the eluent. Therefore, it is suitable for LC/MS analysis of oligonucleotides.
Sample: Synthesized oligo-DNAs (crude), 1 μL
- 1.20mer, ATACCGATTAAGCGAAGTTT
- 2.20mer, ATACCGATTAAGCGAATTTT
- 3.20mer, ATACCGATTAAGCTAATTTT
- 4.20mer, ATACCGATTAATCTAATTTT
- Column
- :Shodex HILICpak VN-50 2D (2.0 mm I.D. x 150 mm)
- Eluent
- :(A) 50 mM HCOONH4 aq./(B) CH3CN
Linear gradient;
(B %) 62 to 56 % (0 to 10 min), 56 % (10 to 20 min), 56 to 62 % (20 to 20.01 min), 62 % (20.01 to 25 min) - Flow rate
- :0.2 mL/min
- Detector
- :UV (260 nm) (small cell volume), ESI-MS (SIM Negative)
- Column temp.
- :60 ℃
Sample Name Index
Operation Manual / Certificate of Analysis
Operation Manuals and Certificate of Analysis / Inspection Certificate for the following products can be downloaded here.
Product Name Index
Applications
- High Sensitive Analysis of Synthetic Oligo-DNAs by LC/MS (Comparison between VN-50 1D and VN-50 2D)
- LC/UV/MS Analysis of Oligo-DNA (VN-50 2D)
- Analysis of Phosphorothioated Oligo-DNA (VN-50 2D)
- Analysis of Oligo-DNAs (VN-50 2D)
- Analysis of Various Oligonucleotides by LC/UV/MS (VN-50 2D)
- Analysis of 2'-OMe and 2'-MOE Modified Phosphorothioated Oligo-RNA by LC/UV/MS (VN-50 2D)
- Analysis of 10 - 50mer Oligo-DNAs (VN-50 2D)
- Analysis of Oligonucleotides and Their Impurities (1) Truncated Oligonucleotides (VN-50 2D)
